In a small attempt at blowing away the cobwebs in this website, I have updated my Publications page. I switched from manual updates (*shudder*) to the Papercite plug-in, which updates from a bibtex file stored in my Dropbox. I am in the process of adding PDF files for every single publication, and experiencing the joys of wading through publisher regulations to check what their conditions are for sharing post-prints.
In this regard, SHERPA/RoMEO has been very helpful. You simply look for a specific journal and see what they allow. For example, the Computers & Graphics journal published by Elsevier places a 24-month embargo on putting your post-print on open access repositories and institutional pages, but does allow author’s to put the accepted manuscript on their personal website or blog immediately. Yay for having a blog!
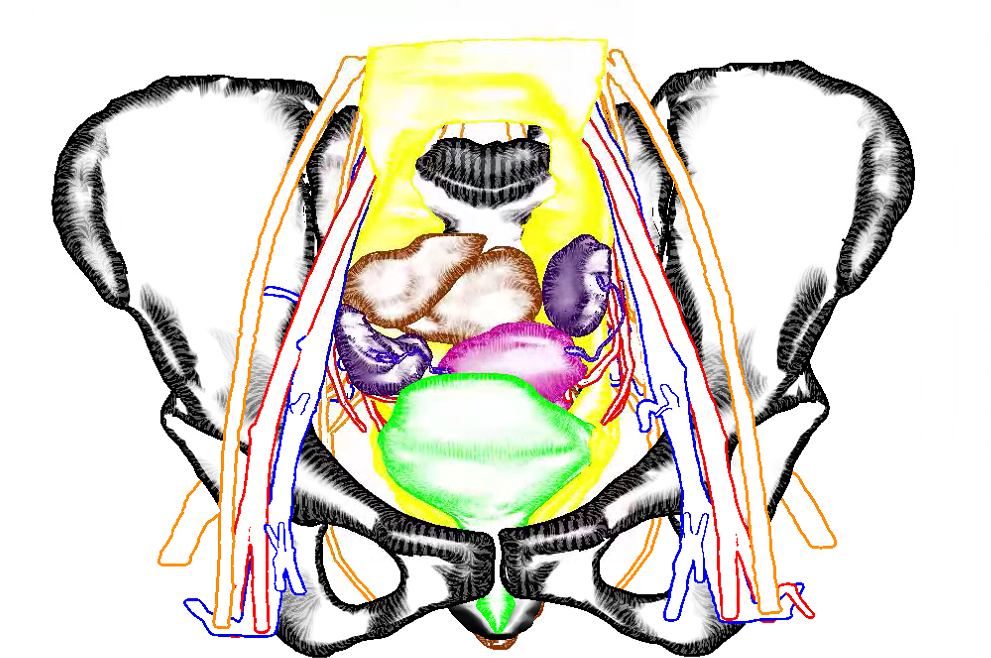
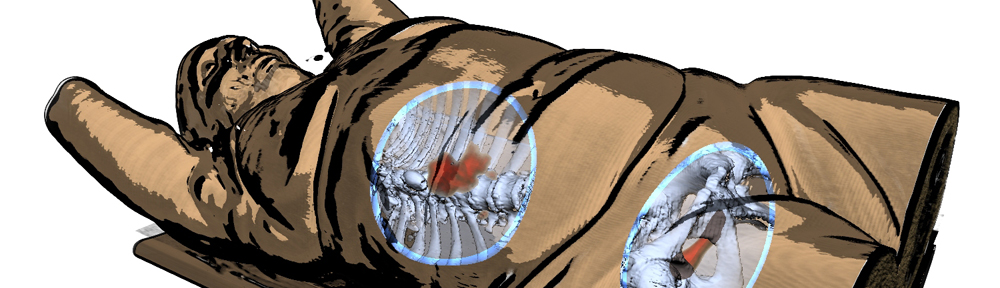
So here is a blog-exclusive, the PDF of our accepted paper ‘Real-time Field Aligned Stripe Patterns'[1] is now hosted on this very website! Another recent publication that I may have failed to mention here so far is our Survey on Multimodal Medical Visualization[2].
[Bibtex]
@article{lichtenberg_real-time_2018,
title = {Real-time field aligned stripe patterns},
volume = {74},
issn = {0097-8493},
url = {http://noeskasmit.com/wp-content/uploads/2018/08/lichtenberg_2018.pdf},
doi = {10.1016/j.cag.2018.04.008},
abstract = {In this paper, we present a parameterization technique that can be applied to surface meshes in real-time without time-consuming preprocessing steps. The parameterization is suitable for the display of (un-)oriented patterns and texture patches, and to sample a surface in a periodic fashion. The method is inspired by existing work that solves a global optimization problem to generate a continuous stripe pattern on the surface, from which texture coordinates can be derived. We propose a local optimization approach that is suitable for parallel execution on the GPU, which drastically reduces computation time. With this, we achieve on-the-fly texturing of 3D, medium-sized (up to 70 k vertices) surface meshes. The algorithm takes a tangent vector field as input and aligns the texture coordinates to it. Our technique achieves real-time parameterization of the surface meshes by employing a parallelizable local search algorithm that converges to a local minimum in a few iterations. The calculation in real-time allows for live parameter updates and determination of varying texture coordinates. Furthermore, the method can handle non-manifold meshes. The technique is useful in various applications, e.g., biomedical visualization and flow visualization. We highlight our method{\textquoteright}s potential by providing usage scenarios for several applications.},
urldate = {2018-07-29},
journal = {{Computers \& Graphics}},
author = {Lichtenberg, Nils and Smit, Noeska and Hansen, Christian and Lawonn, Kai},
month = aug,
year = {2018},
keywords = {Computational geometry,, Computer graphics, Parameterization, Visualization},
pages = {137--149},
}[Bibtex]
@article{lawonn_survey_2017,
title = {A {Survey} on {Multimodal} {Medical} {Data} {Visualization}},
doi = {10.1111/cgf.13306},
journal = {{Computer Graphics Forum}},
author = {Lawonn, Kai and Smit, Noeska and B{\"u}hler, Katja and Preim, Bernhard},
abstract = {Multimodal data of the complex human anatomy contain a wealth of information. To visualize and explore such data, techniques
for emphasizing important structures and controlling visibility are essential. Such fused overview visualizations guide
physicians to suspicious regions to be analyzed in detail, e.g. with slice-based viewing. We give an overview of state of the art
in multimodal medical data visualization techniques. Multimodal medical data consists of multiple scans of the same subject
using various acquisition methods, often combining multiple complimentary types of information. Three-dimensional visualization
techniques for multimodal medical data can be used in diagnosis, treatment planning, doctor-patient communication
as well as interdisciplinary communication. Over the years, multiple techniques have been developed in order to cope with
the various associated challenges and present the relevant information from multiple sources in an insightful way. We present
an overview of these techniques and analyze the specific challenges that arise in multimodal data visualization and how recent
works aimed to solve these, often using smart visibility techniques. We provide a taxonomy of these multimodal visualization applications
based on the modalities used and the visualization techniques employed. Additionally, we identify unsolved problems
as potential future research directions.
},
month = oct,
year = {2017},
url = {https://vis.uib.no/wp-content/papercite-data/pdfs/LawonnSmit-2017-MULTI.pdf}
}